Speaker: Marinka Zitnik (Harvard)
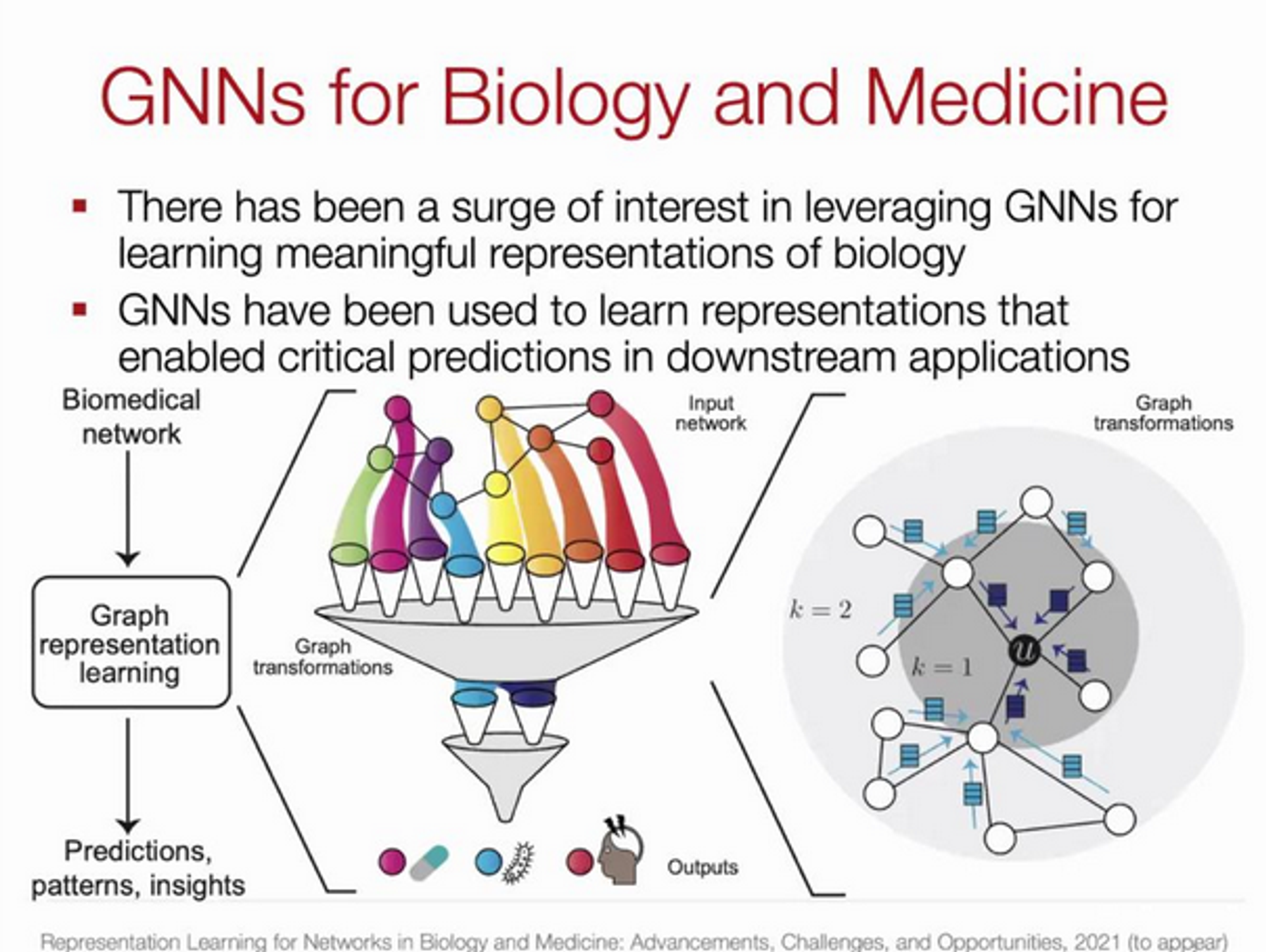
What makes graph representation techniques well suited for the analysis of high-dimensional interconnected medical data?
→ Biological systems are interconnected at different scales:
- e.g. RNA-proteins-compounds-disease
- Patient networks
- Hierarchies of cell systems
- Disease pathways
- Biomenical knowledge graphs
- Gene interaction networks
- Cell-cell-similarity netowrks
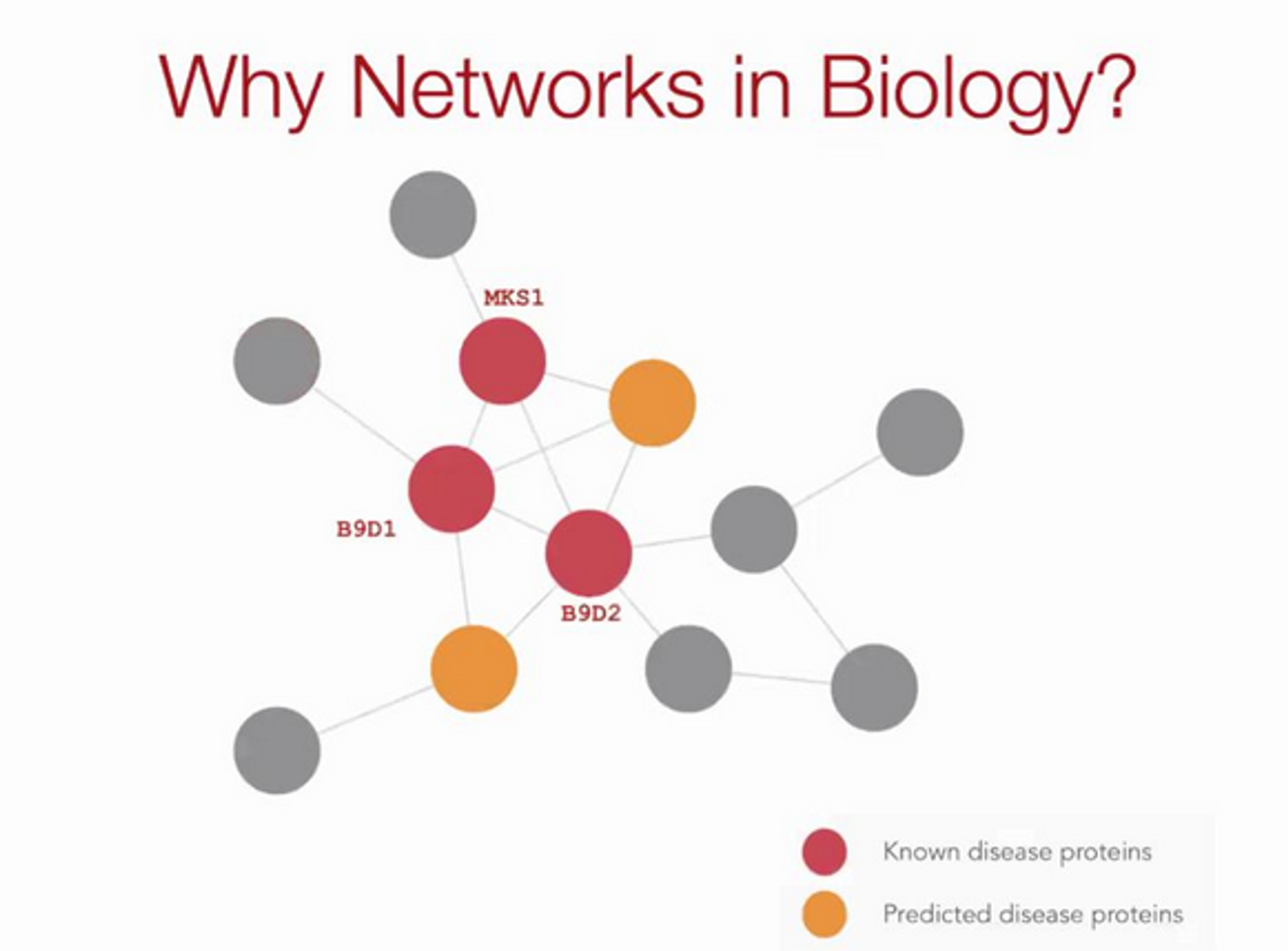
Meta learning for graphs
Never-before-seen disease → we want to repurpose existing drugs as approval of new drugs time-costly.
Why is finding treatments for new disease challenging?
- Generalising to new phenomena is hard
- Prevailing GNN methods require abundant label information
- However, labeled examples are scarce
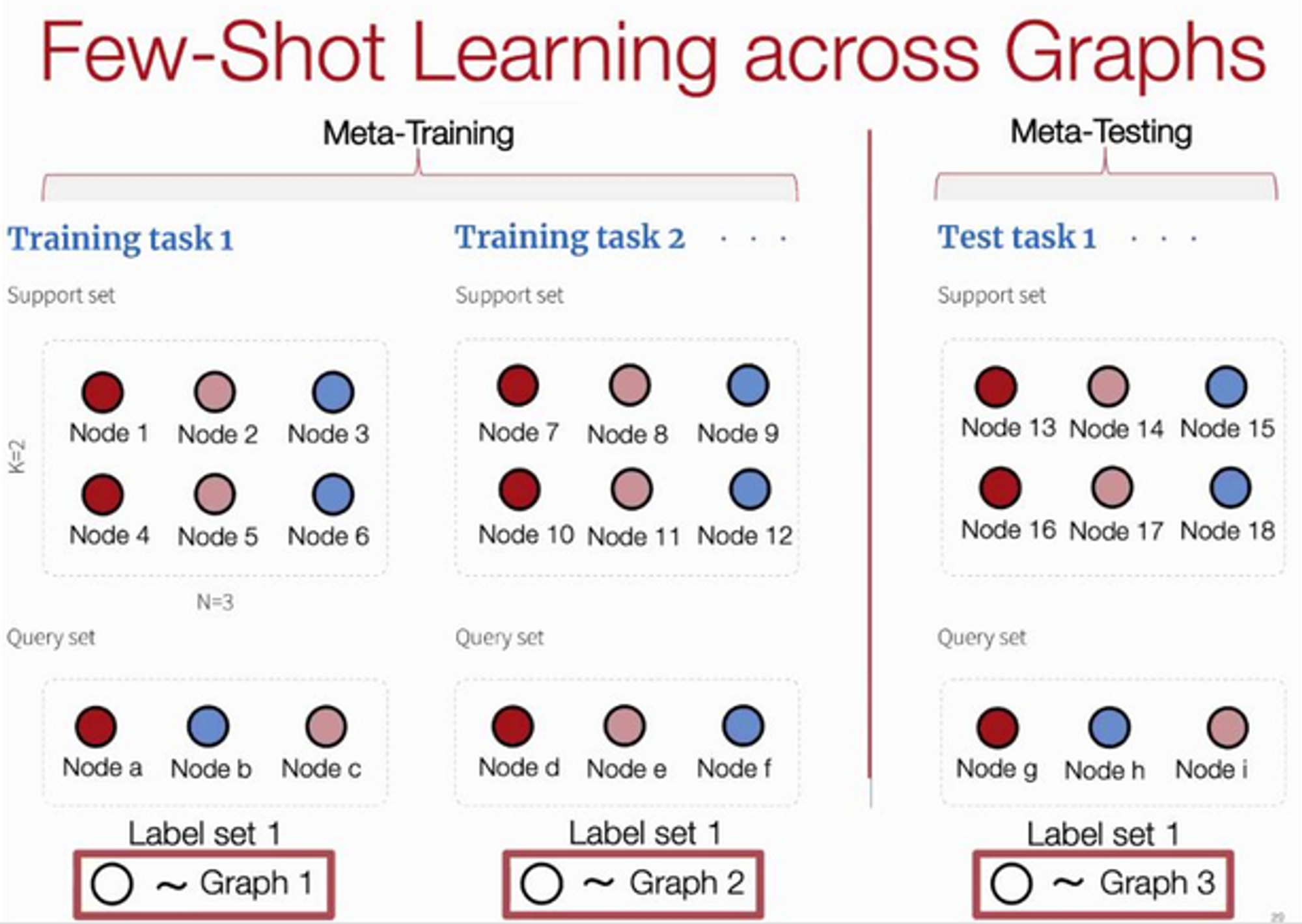
The question is then: how can we design powerful meta-learners which can transfer learning from one labeled example to others? How to make predictions on a new graph when we only have an handful of labels?
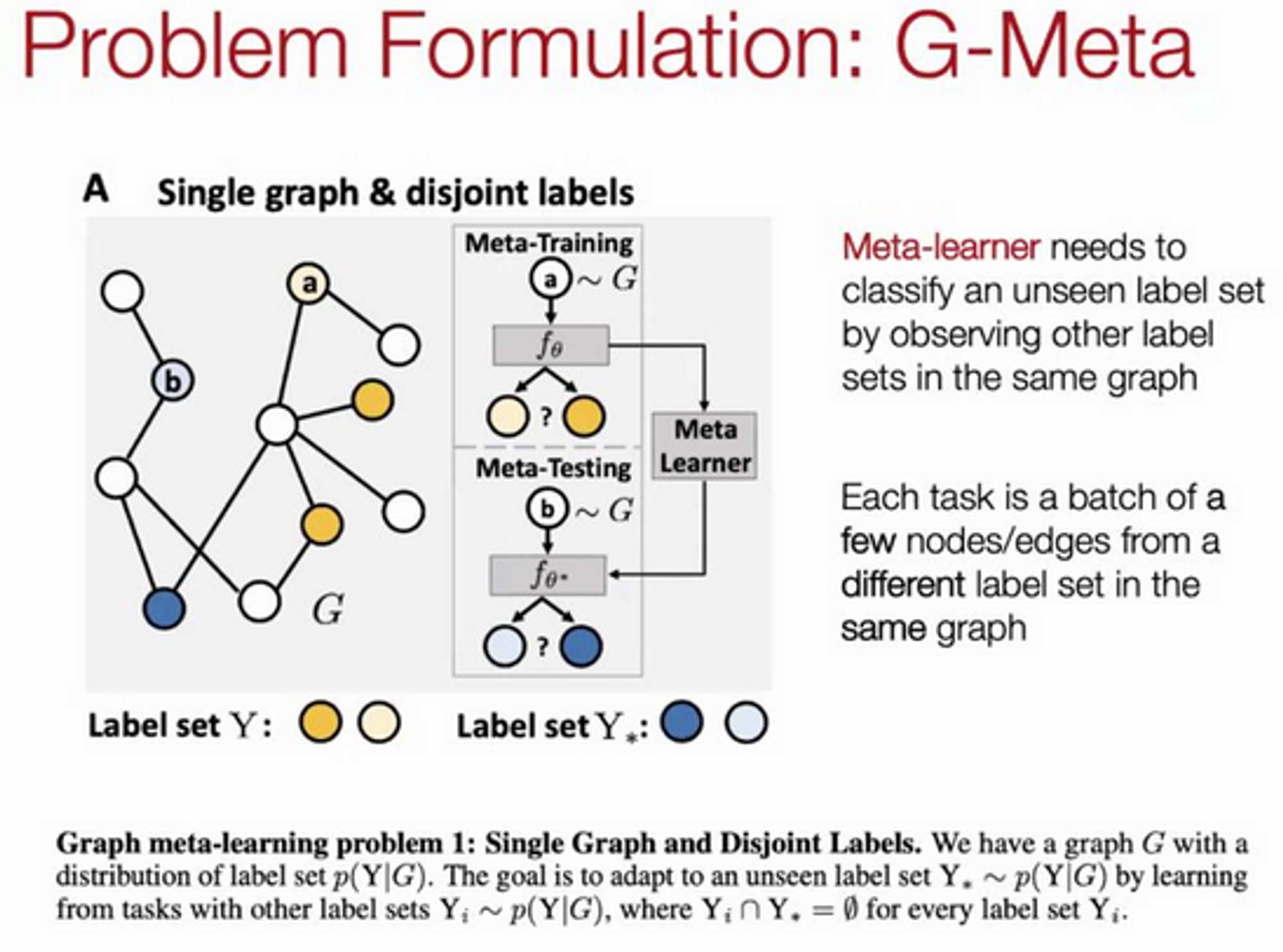
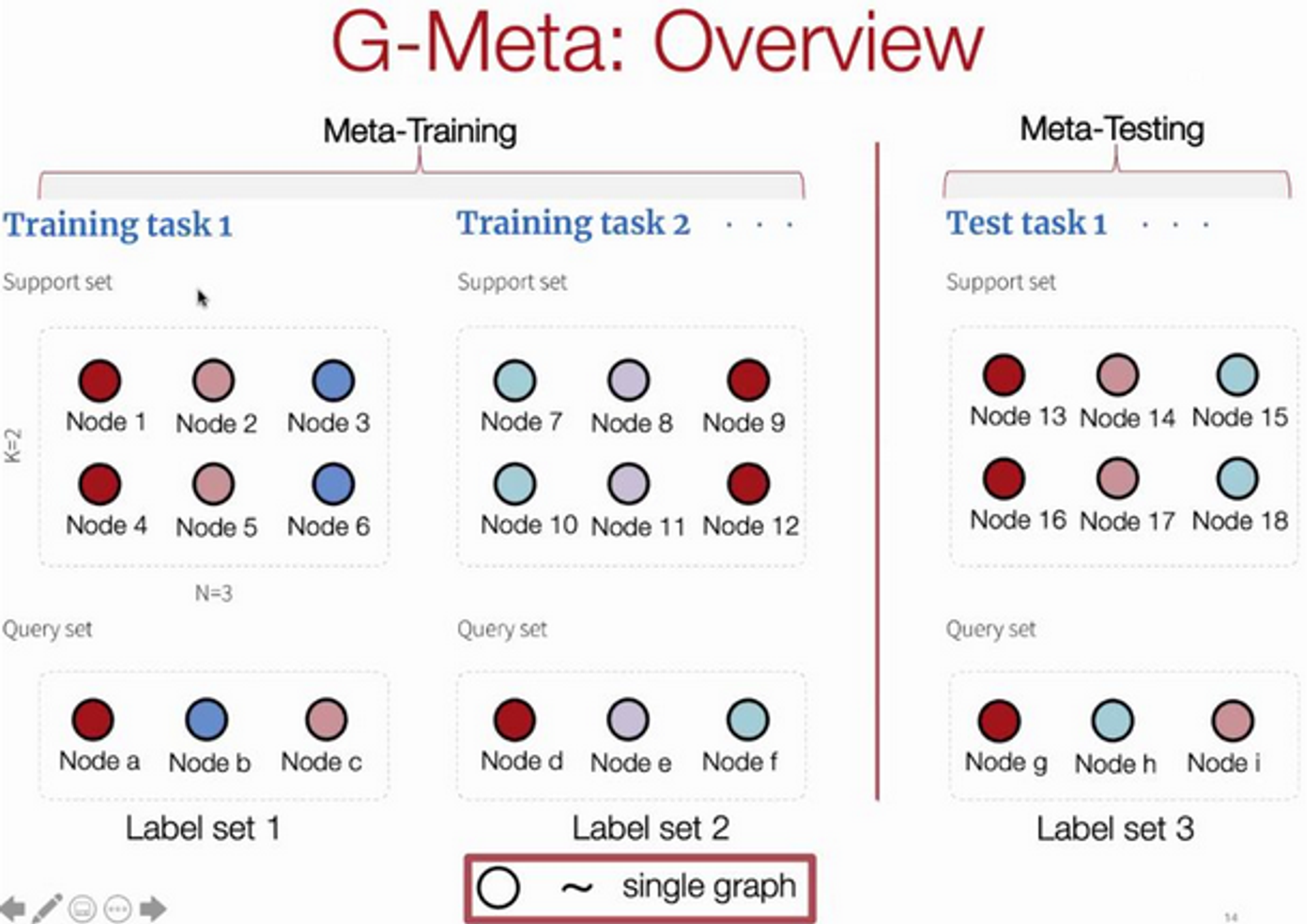
Key idea: local subgraphs - consider a distribution over subgraphs as the distribution over tasks from which a global set of parameters are learned.
Use this strategy to do link prediction.
Why are subgraphs useful:

When labels are scarse, label propagation is not sufficient → here structure similarity is more useful.
G-Meta learns a metric to classify query subgraphs using the closest point from the support set.
COVID-19 Drug Repurposing
COVID-19 Repurposing Dataset

What human proteins does the virus bind to? Interactions between human and protein graphs.
How to represent COVID-19? Network neighbourhood of human PPI network targeted by virus.
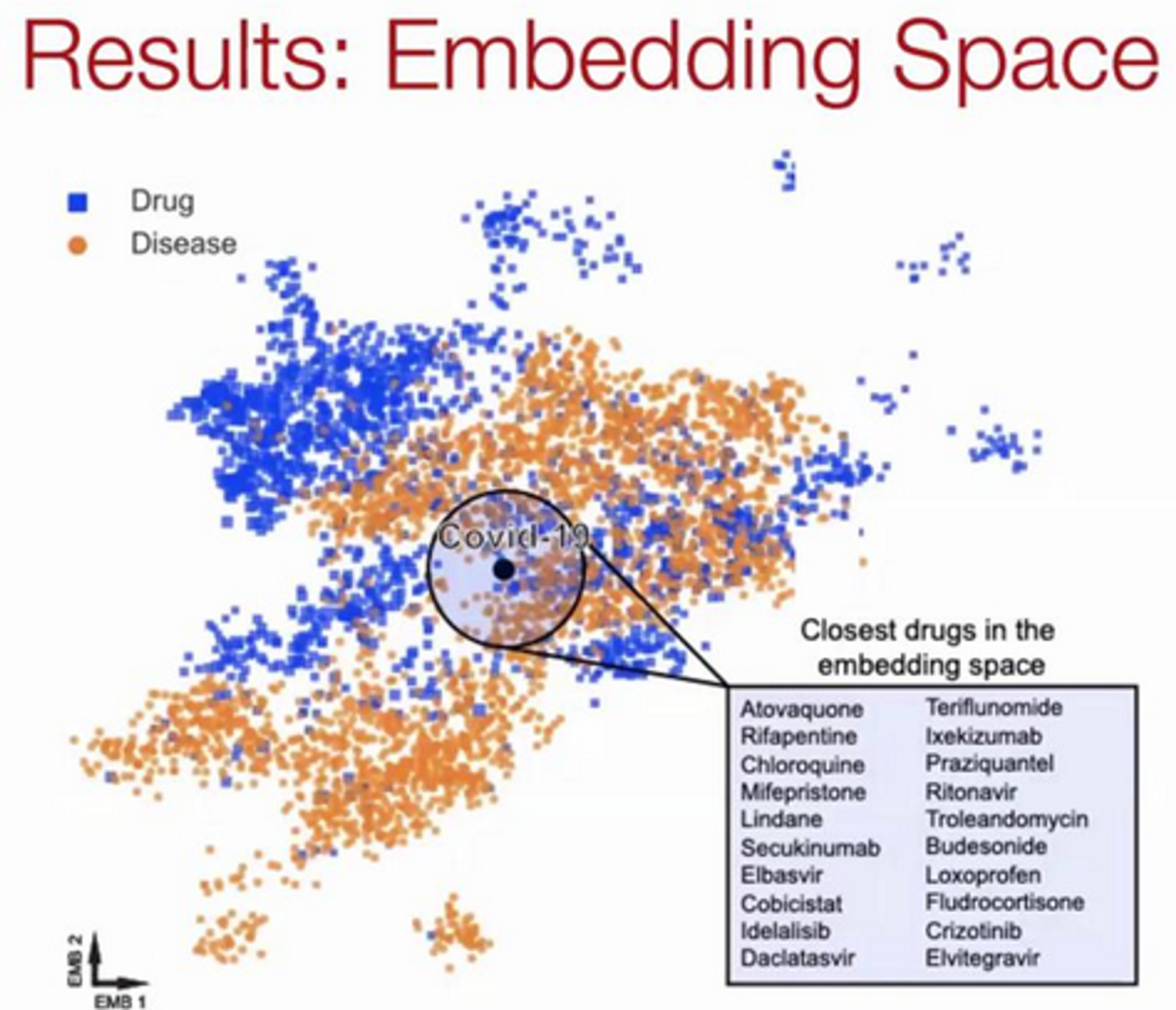
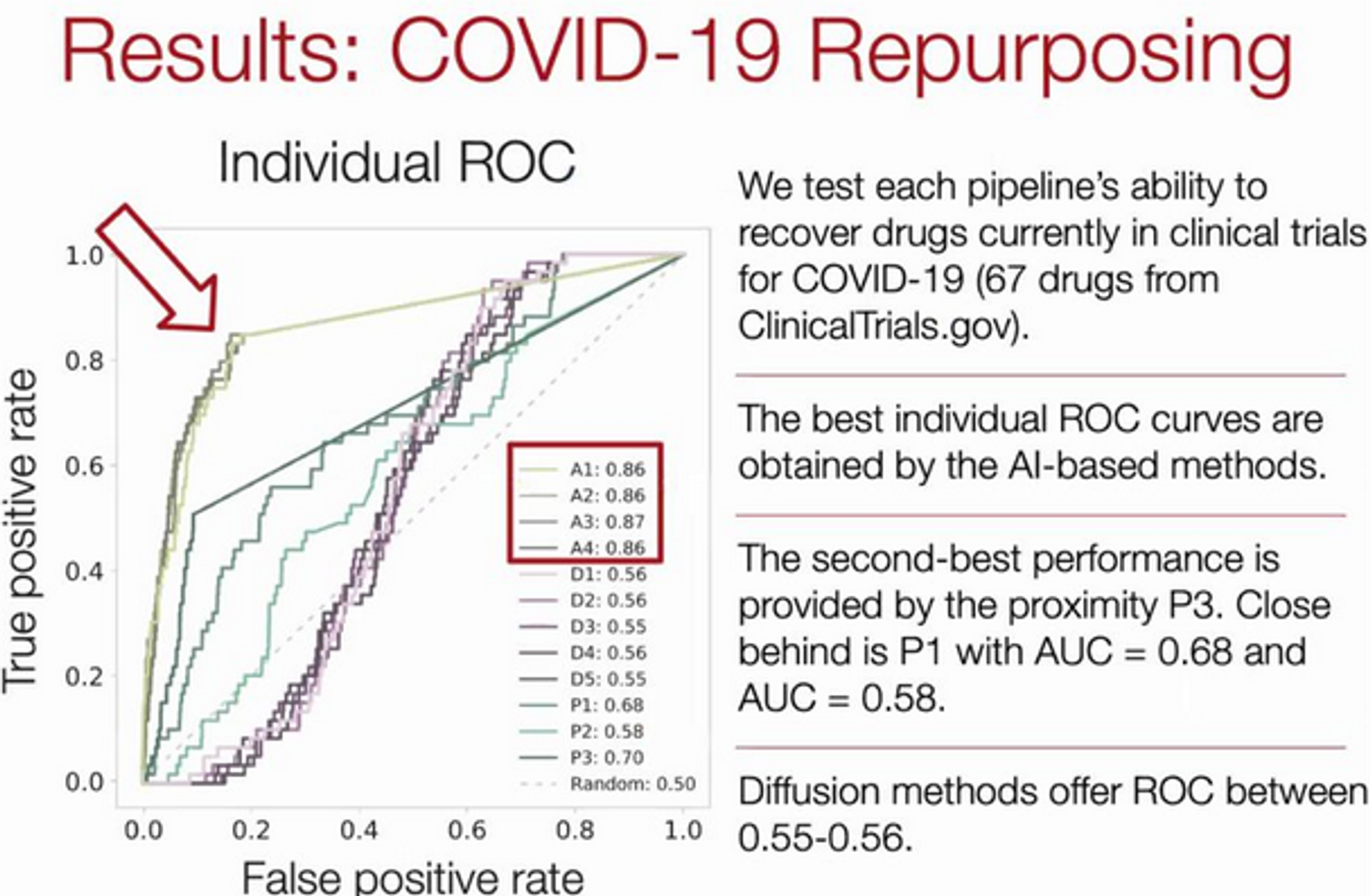

Results:

Interesting Finding: 76/77 drugs that successfully reduced viral infections do not directly bind proteins targeted by COVID.
These drugs rely on network-based actions that can's be identified by traditional docking-based strategies.
Key ML Lessons
- Domain scientists without AI expertise still need a way to interact with AI systems and need to be able to feedback in the ML loop.
- Zero-shot learning: generalising to new graphs is hard.
Between Organisms
Additional idea: how can we leverage this different type of graph transfer learning? Can we learn for humans from what we know on other organisms? G-Meta can also be used here.
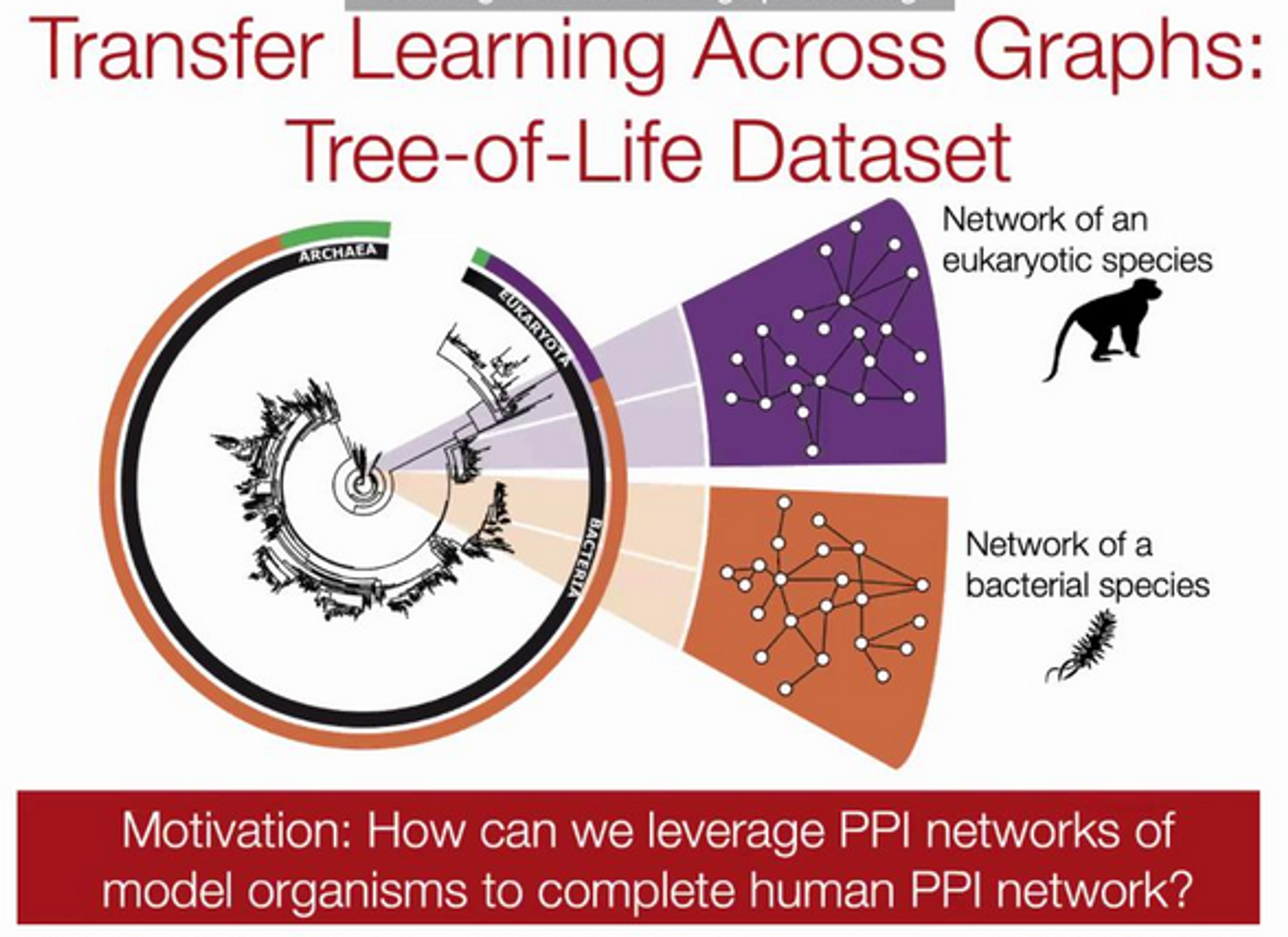
Here G-META is 29.9% over previous works and can scale to large graphs - 100x increase in graph size.
Therapeutics Data Commons
First unified framework to systematically evaluate ML across range of therapeutics. Creates new opportunities for graph-learning to be applied.
