Speaker: Jonathan Godwin (DeepMind)
🦒 Jraph
A high-performance GNN library.
Built in JAX, powered by XLA.
Easily forkable.
(More) easily sharded for ⬆️ models (i.e. large graphs).
JAX Overview
Key features:
- Normal numpy + autodiff. E.g.. twice differentiate ... just by calling
grad()twice!
- Built-in XLA (recall: XLA (Accelerated Linear Algebra) = domain-specific compiler for linear algebra) - one-liner for compilation
- Easy SPMD parallelism - see
pmapfunction, which gives automatic cross-device parallelism
What do we optimise for?
Interested in wide-range of applications - focus is on the most general Graph Nets framework (i.e. the Battaglia paper)

Examples given of fluid/materials & mesh physics prediction papers. I recall that the former used the (old?) deepmind graph nets library. Assume the latter paper uses Jraph?
Jraph Design
🧠 What are the key design criteria?
Functional: don't want to have explicit state - we want to outline a compute pattern
➡️ very much in favour of this approach; abstracting state from flow not only computational gain, but also much more intuitive conceptually
Forkable: copy-pastable code; limit the number of abstractions
➡️ I really like this criterion, often overlooked
Padding & Masking: XLA has a static shape requirement for tensors, which filters down into JAX and Jraph - so we have to pad graphs if they're dynamically shaped and we need masking to make sure we don't accidentally pass messages "from padding"
Easy to use Jax parallelism for new models
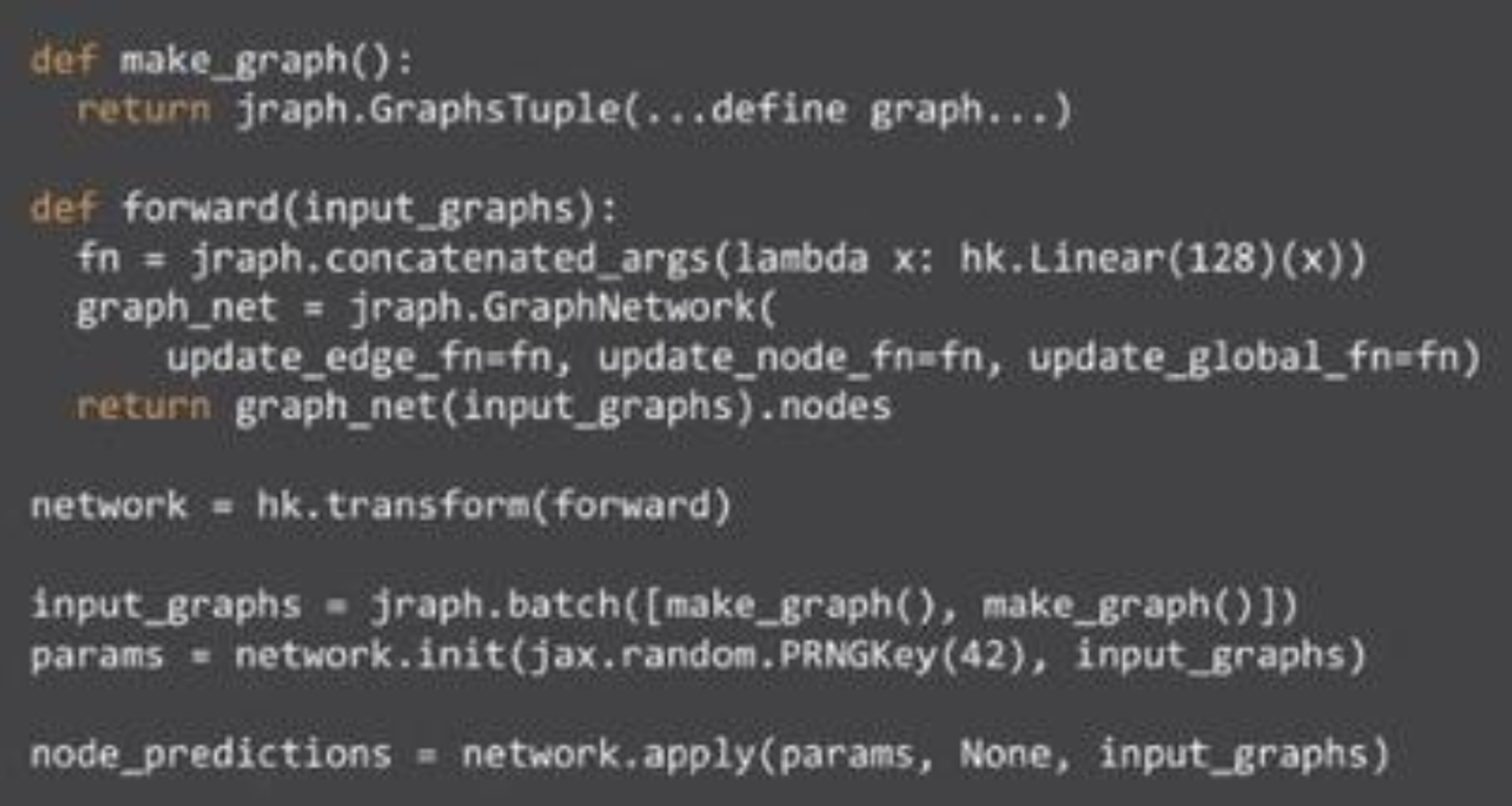
Functional GNNs
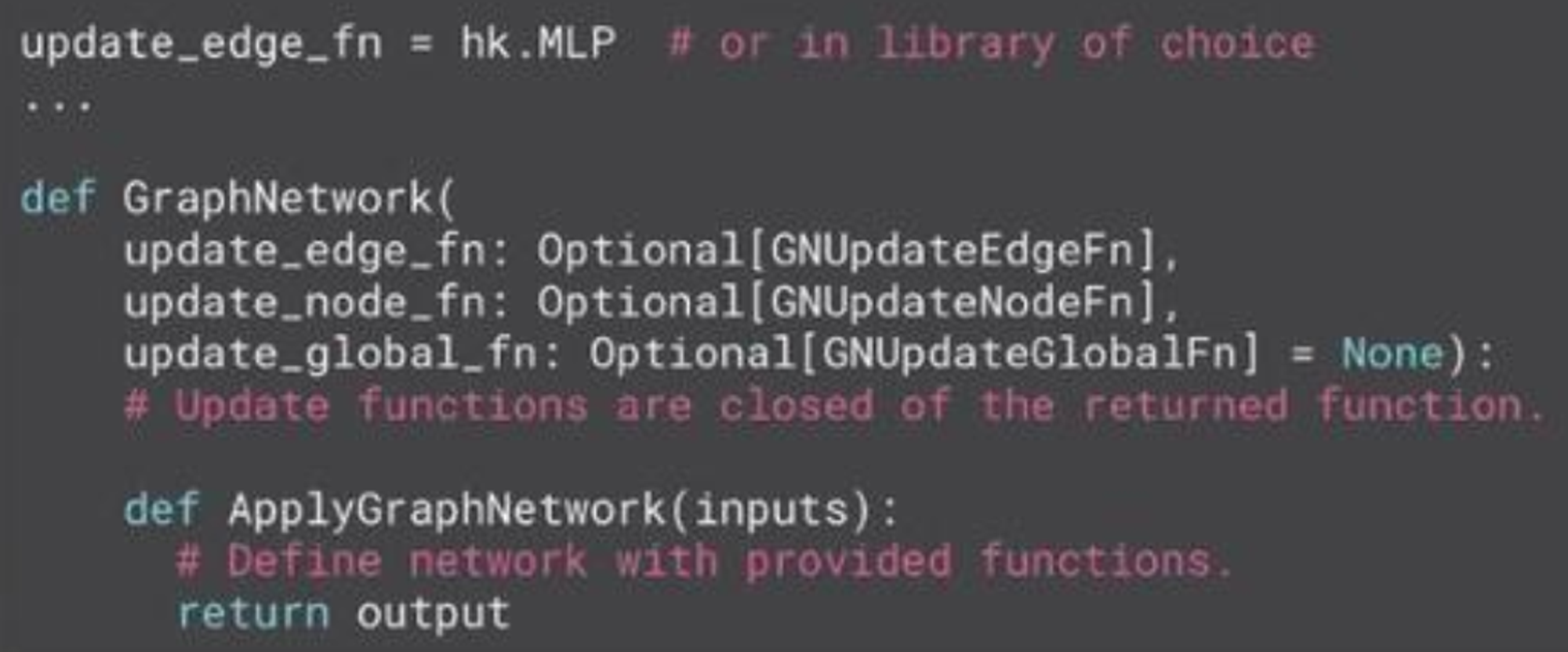
Key idea is that GNNs as functions that define a computation pattern (if you want to use state → Haiku or Flax).
Update functions are how learned params are injected. Inserted via a closure during network construction.
Forkable
Simple enough that you could just copy-paste a model and then modify to run your own version, without having to wory about various abstractions.
Static Shape Utilities
Provide tools to deal with necessary padding and masking (XLA req.).
Support For Nests
Can easily make recurrence models work - e.g. LSTM edge-to-node aggregation (pictured), which easily allows LSTM state and node embedding to be handled:

Advanced Features Coming Soon: Massive GNNs
How might you implement a distributed GNN?
E.g. graph has many millions of edges/nodes and can't fit on a single device.
But we know that edge and node updates are highly data parallel!
- Split up edge tensors onto different devices
- Update edges on separate devices
- For node updates, naively: we might send a single edge row between devices (expensive! avoid!)
- Instead, replicate all nodes on all devices and do a partial reduce of edge updates available within a device
- Becore doing a second cross-device sum reduce
This is very simple and very neat - takes us from to and spreads the workload out nicely.
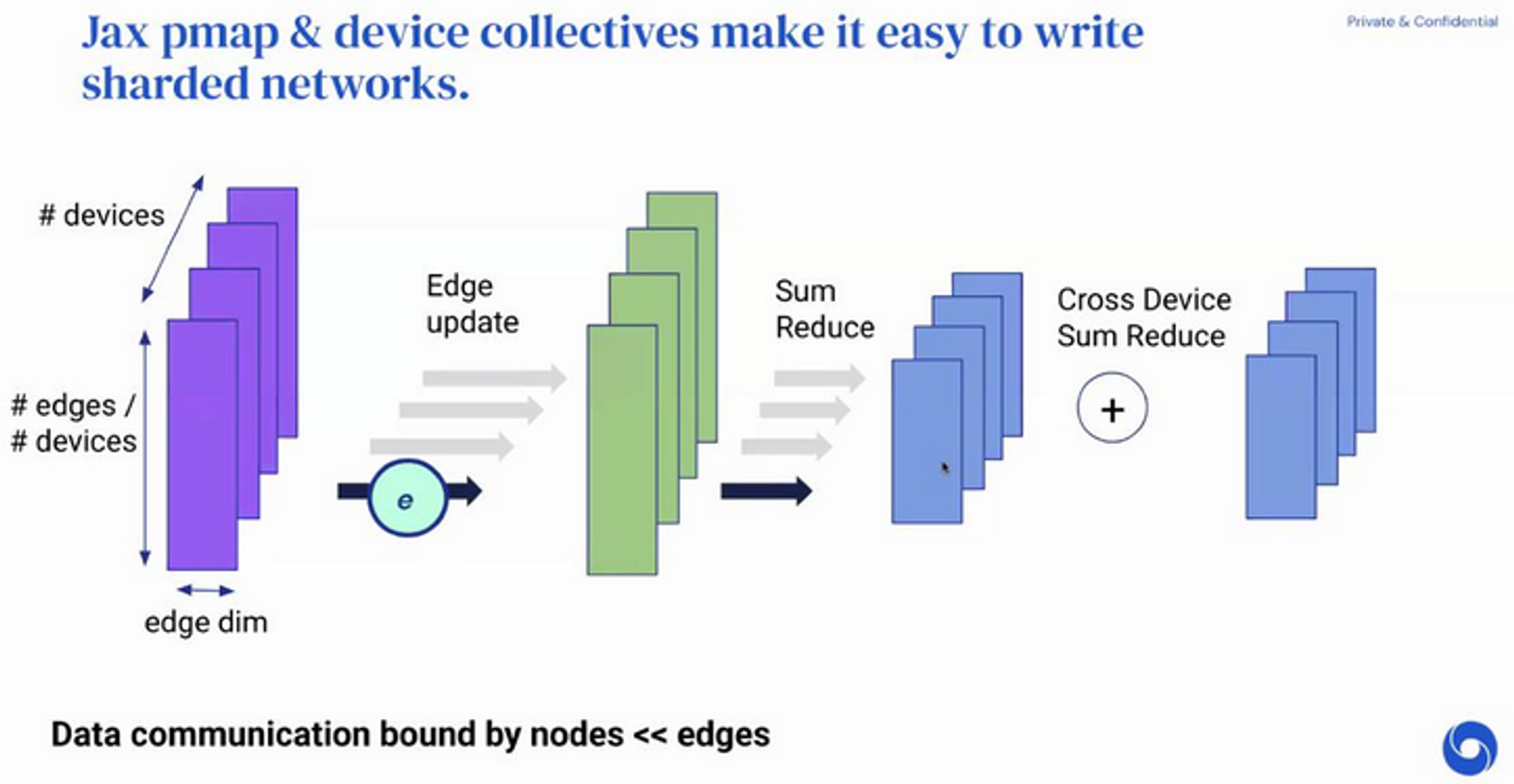
Also, note that these node summations simply send the entire node tensor (no transpose, permutations, etc) improving utilisation of network bandwidth.
This is all based on the assumption that we can fit nodes in mem but not edges.
The
pmap and psum commands in Jax will take care of edge and node updates respectively 🙂Results
(on TPUv1)
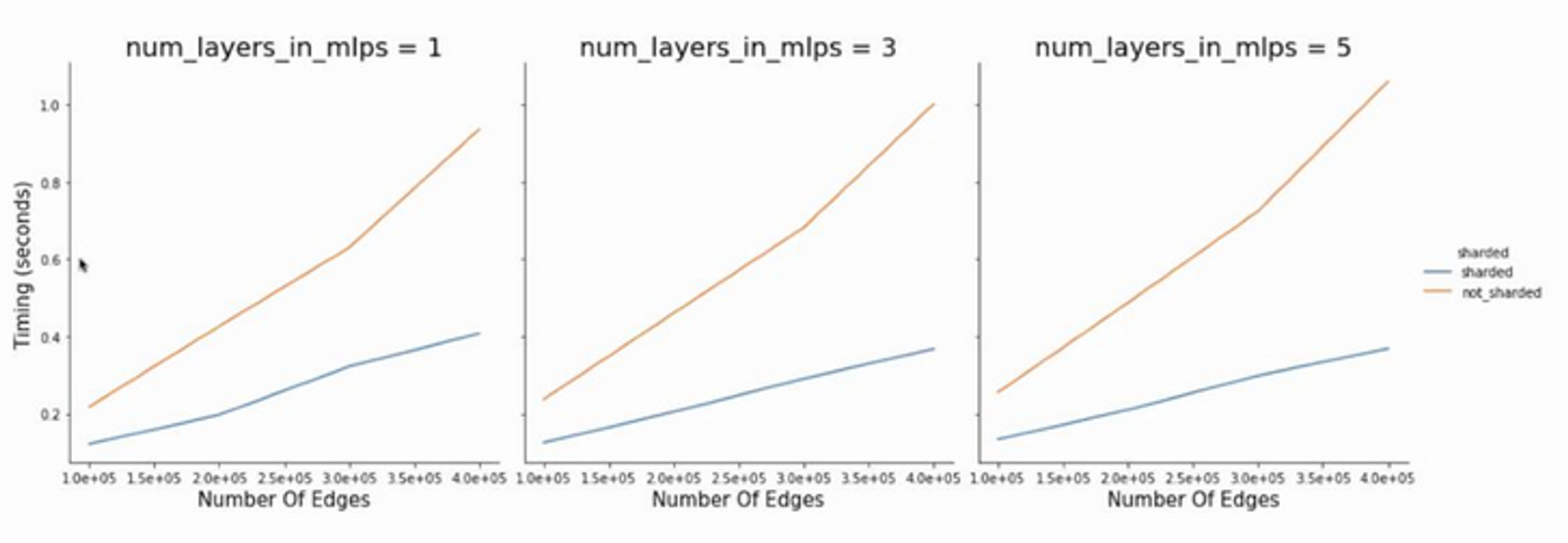
Very large systems can now be trained! On the path to GPT-3 for graph nets (maybe 😁)
Q & A
What are the considerations for data (batch) vs model vs pipeline parallelism?
Depends on the problem at hand. In graph-land(!) we have lots of large graphs, e.g. using large embeddings as well as many nodes/edges, so data-parallelism will be required.
Models themselves tend not to be very deep/have many params, so perhaps model parallelism not useful.
Pipeline model parallelism may be more useful. Benefit: you can send the entire graph across each stage in your pipeline, which will improve bandwidth utilisation.
Padding & masking may introduce overheads for computation, how do we optimise that compared to batching everything in a single graph (what PyTorch geometric would do)?
Can't really get away from if you want to run on TPUs or use XLA.
We would still take the approach of padding everything into a single batch - e.g. multiple graphs in a batch might have different sizes/shapes.
But can utilise a dynamic number of batches.